paired end sequencing advantages
For example with splicing junction events two exons maybe joined together. Paired-end reading improves the ability to identify the relative positions of various reads in the genome making it much more effective than single-end reading in resolving structural rearrangements such as.

Cell Free Fetal Dna Coming In All Sizes And Shapes Chiu 2021 Prenatal Diagnosis Wiley Online Library
Illumina sequencing by synthesis technology supports both single-read and paired-end libraries.

. SOLiD is another massively parallel short-tag sequencing platform introduced in late 2007 by Applied Biosystems. 1 Better mapping results. Response to exercise 6.
Paired-End Reads Alignment to the Reference Sequence Repeats Reference Figure 4. The paired-end PE approach where each molecule is sequenced from both the 5 and 3 ends can double the number of bp per read for the Illumina platform. The advantage Ive seen of paired end sequencing is that in mRNA analysis when you sequence the RNA cDNA and want to map it against the reference genome you end up facing a problem which is that cDNA does not.
8 cycles for the Index 1 i7 Read 8 cycles for the Index 2 i5 Read and 7 nonimaging chemistry-only cycles at the beginning of the i5 Read. This application is called pairwise end sequencing known colloquially as double-barrel shotgun sequencing. NGS analysis Illumina sequencing Benefits of paired end sequencing.
Fast and Accurate Next-Generation Sequencing Results Enabled by Ion Torrent Technology. Because PET represent connectivity between the tags the use of PET in genome re-sequencing has advantages over the use of single reads. Specifically the invention provides cloning and DNA manipulation strategies to isolate the two ends of a large target nucleic acid into a single small DNA construct for rapid cloning.
Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data. Since paired-end reads are more likely to align to a reference the quality of the entire data set. There are several advantages of using dual indexes.
NGS analysis Illumina sequencing Benefits of paired end sequencing. Approximately 100-200 tumors can be sequenced at a resolution greater than 150kb when compared to sequencing an entire genome. There are several benefits of sequencing your DNA.
With PE reads you know roughly the range of the distance of two reads. The advantage Ive seen of paired end sequencing is that in mRNA analysis when you sequence the RNA cDNA and want to map it against the reference genome you end up facing a problem which is that cDNA does not contain the introns. Another supposed advantage is that it leads to more accurate reads because if say Read 1 see picture below maps to two different regions of the genome Read 2 can be used to help determine which one of the two regions makes more sense.
Paired-End Sequencing and Alignment Paired-end sequencing enables both ends of the DNA fragment to be sequenced. Intolerance to certain types of foods 5. For paired-end flow cells dual indexing introduces 23 additional cycles of sequencing.
I am quite new with the terminology. This platform was adapted from the polony sequencing method Shendure et al. Disease risk prediction 2.
The shorter Illumina reads may reduce phylogenetic resolution both in terms of picking operational taxonomic units OTUs and determining evolutionary distances between OTUs. Status of carrying a disease linked allele that could be passed on to your kids 4. I am new with the Illumina sequencing and I got back the sequencing data.
One of the advantages of paired end sequencing over single end is that it doubles the amount of data. SOLiD sequencing is a next gen DNA sequencing method developed by Applied Biosystems. Bearing the limits of short tags in mind the current version of SOLiD is designed.
Benefits of paired end sequencing. Because the distance between each paired read is known alignment algorithms can use this information to map the reads over repetitive regions more. Requires the same amount of DNA as single-read genomic DNA or cDNA sequencing.
By clicking Accept All you consent to the use of ALL the cookies. Can you explain to me what it means that a. The present invention provides for a method of preparing a target nucleic acid fragments to produce a smaller nucleic acid which comprises the two ends of the target nucleic acid.
You can find out your 1. Furthermore Sanger sequencing is analogical while next-generation sequencing is digital allowing the detection of. Yes paired-end sequencing can be done with single.
We use cookies on our website to give you the most relevant experience by remembering your preferences and repeat visits. This may help you when you map the reads back to a reference genome. Yes paired-end sequencing can be done with single.
Response to drugs 3. PE sequencing has the following advantages over SE. For longer DNA fragments paired-end sequencing has to be done through making PET libraries first.
Anchoring one half of the pair uniquely to a single location in the genome allows mapping of the other half that is. The advantage Ive seen of paired end sequencing is that in mRNA analysis when you sequence the RNA cDNA and want to map it against the reference genome you end up facing a problem which is that cDNA does not contain the introns. Paired-end sequencing facilitates detection of genomic rearrangements and repetitive sequence elements as well as gene fusions and novel transcripts.
Because ESP only looks at short paired-end sequences it has the advantage of providing useful information genome-wide without the need for large-scale sequencing. Therefore direct merging of the paired-end reads can prevent potential removal of informative reads that do not comply by the trimming tools strict checks. FLASH to be an efficient tool in conserving reads while carrying out quality trimming in moderation.
Overall our results show that merging paired-end reads of eDNA data before trimming. Ad Gene Expression Profiling Chromosome Counting Epigenetic Changes Molecular Analysis. In paired-end reading it starts at one read finishes this direction at the specified read length and then starts another round of reading from the opposite end of the fragment.
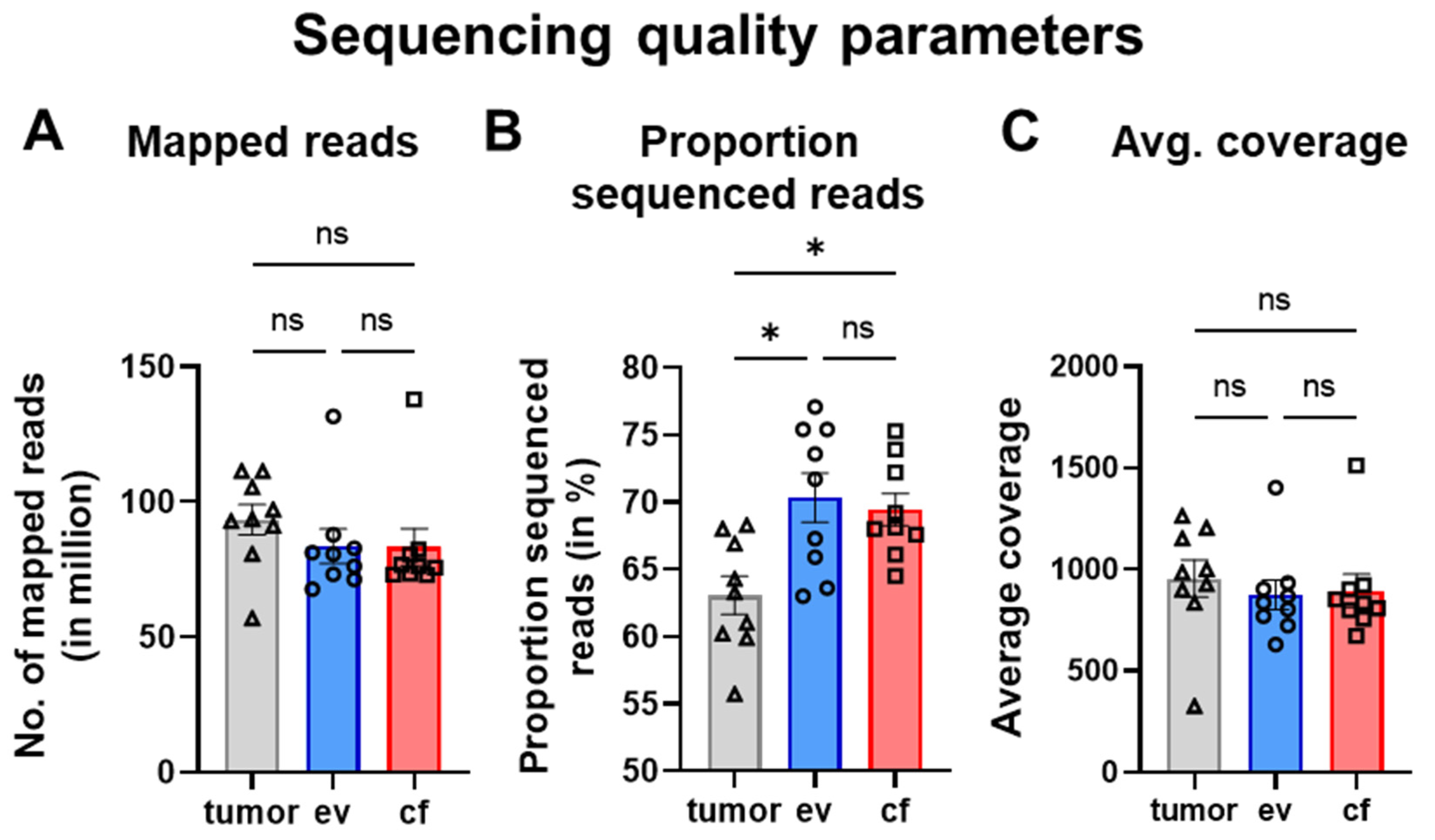
Cancers Free Full Text Comparative Panel Sequencing Of Dna Variants In Cf Ev And Tumordna For Pancreatic Ductal Adenocarcinoma Patients Html

The Variables For Ngs Experiments Coverage Read Length Multiplexing

What Is Mate Pair Sequencing For
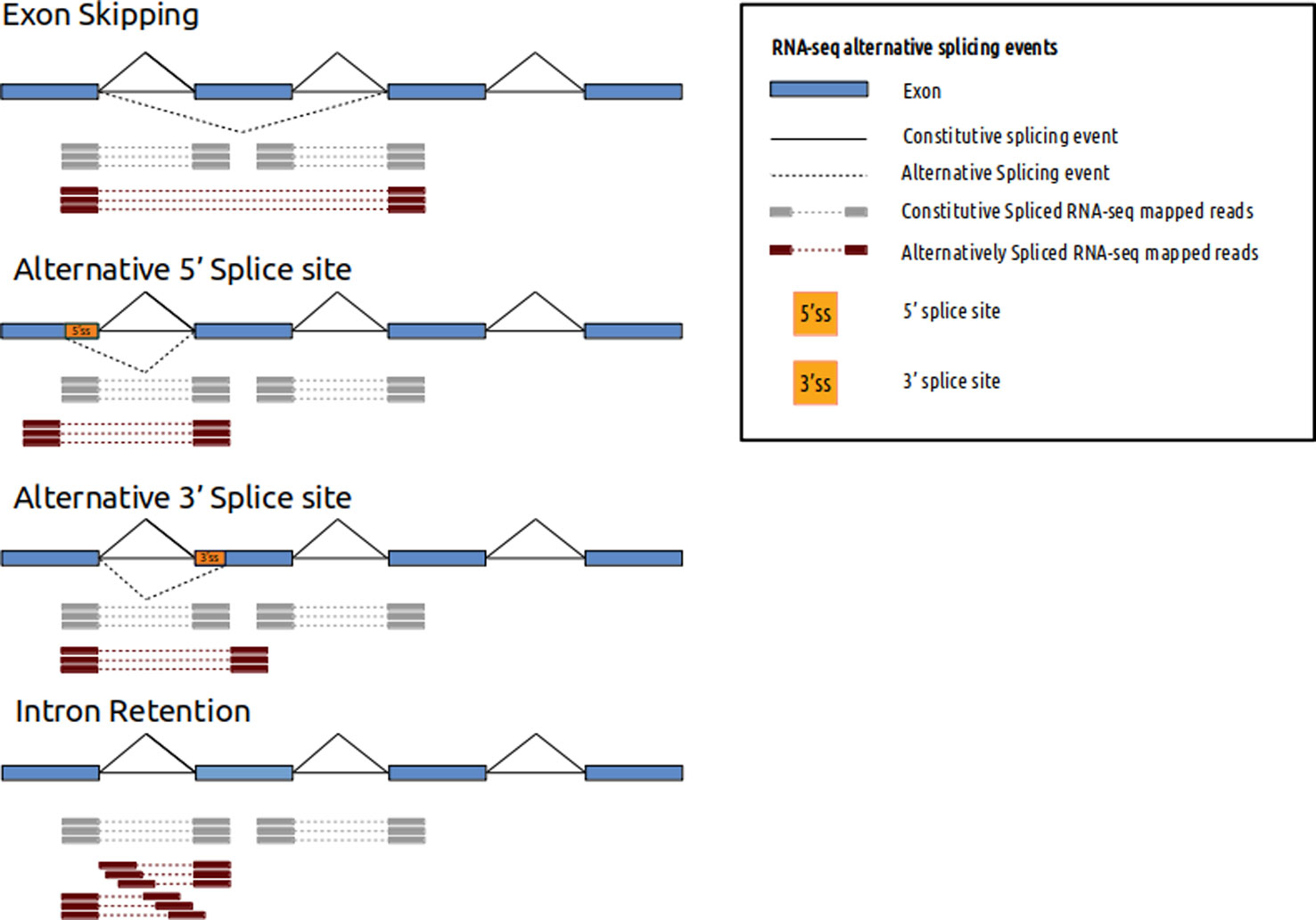
Single End Sequencing Versus Paired End

Single End And Paired End De Novo Assembly Comparisons A K Mer Length Download Scientific Diagram
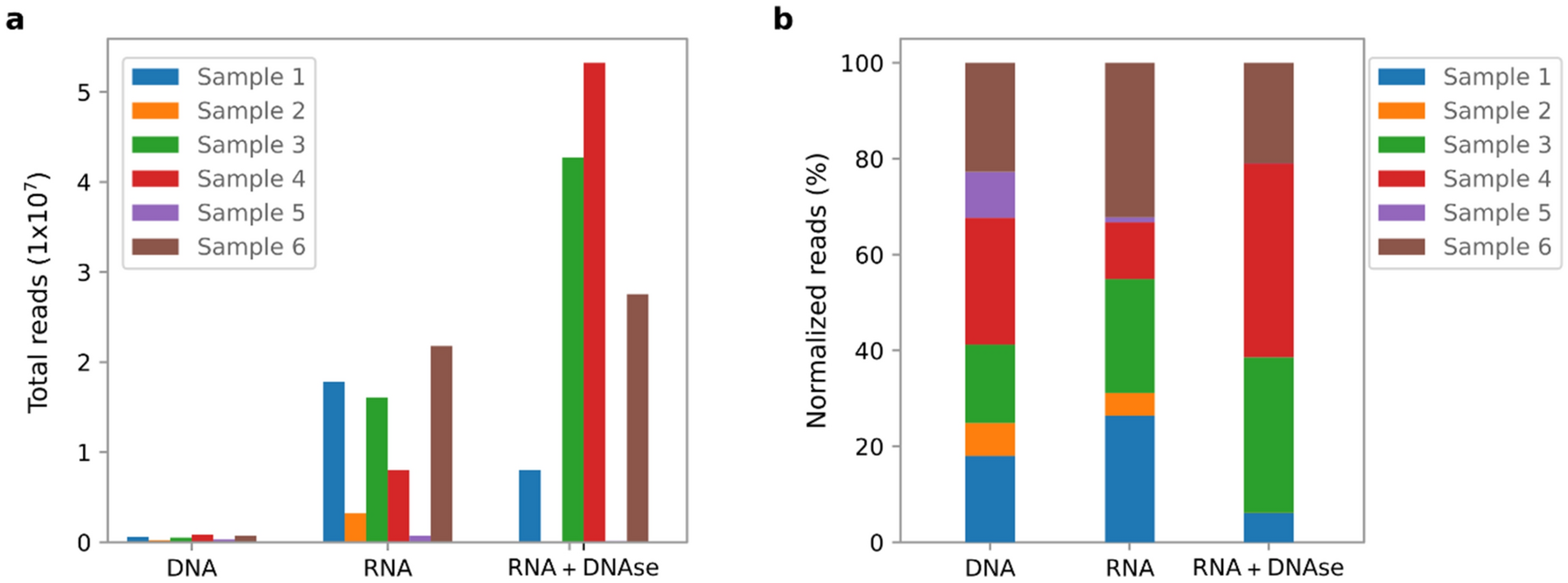
Comparison Of Dna And Rna Sequencing Of Total Nucleic Acids From Human Cervix For Metagenomics Scientific Reports

What Are Paired End Reads The Sequencing Center

Illustrations Of Paired End Sequencing A Illustrates Two Strands Of Download Scientific Diagram
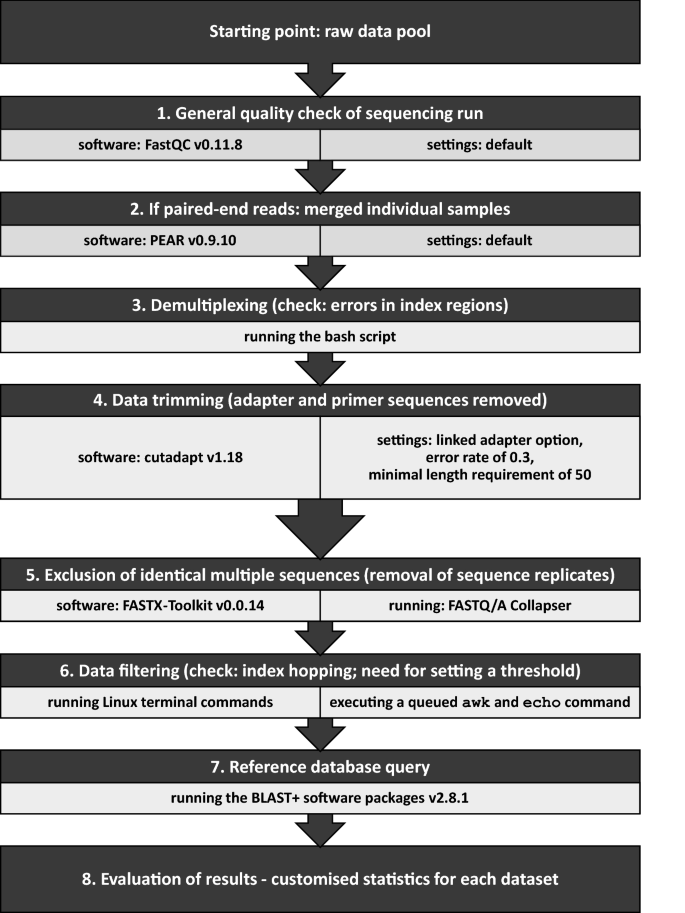
Handling Of Targeted Amplicon Sequencing Data Focusing On Index Hopping And Demultiplexing Using A Nested Metabarcoding Approach In Ecology Scientific Reports
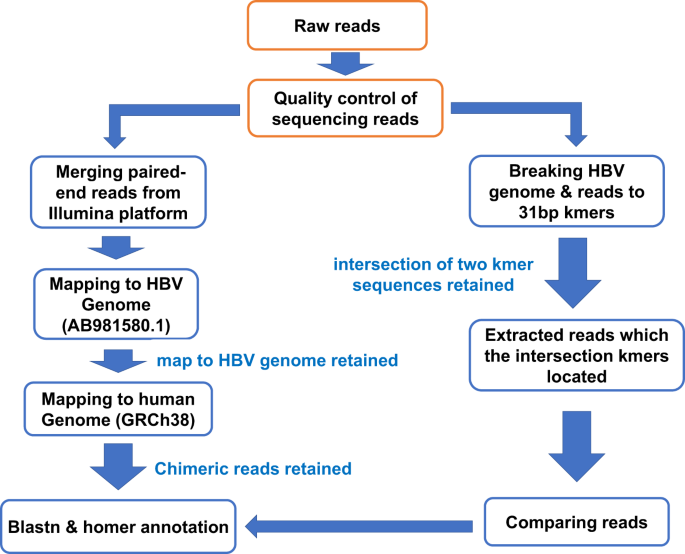
Long Read Sequencing Reveals The Structural Complexity Of Genomic Integration Of Hbv Dna In Hepatocellular Carcinoma Npj Genomic Medicine

Illumina Paired End Sequencing Youtube
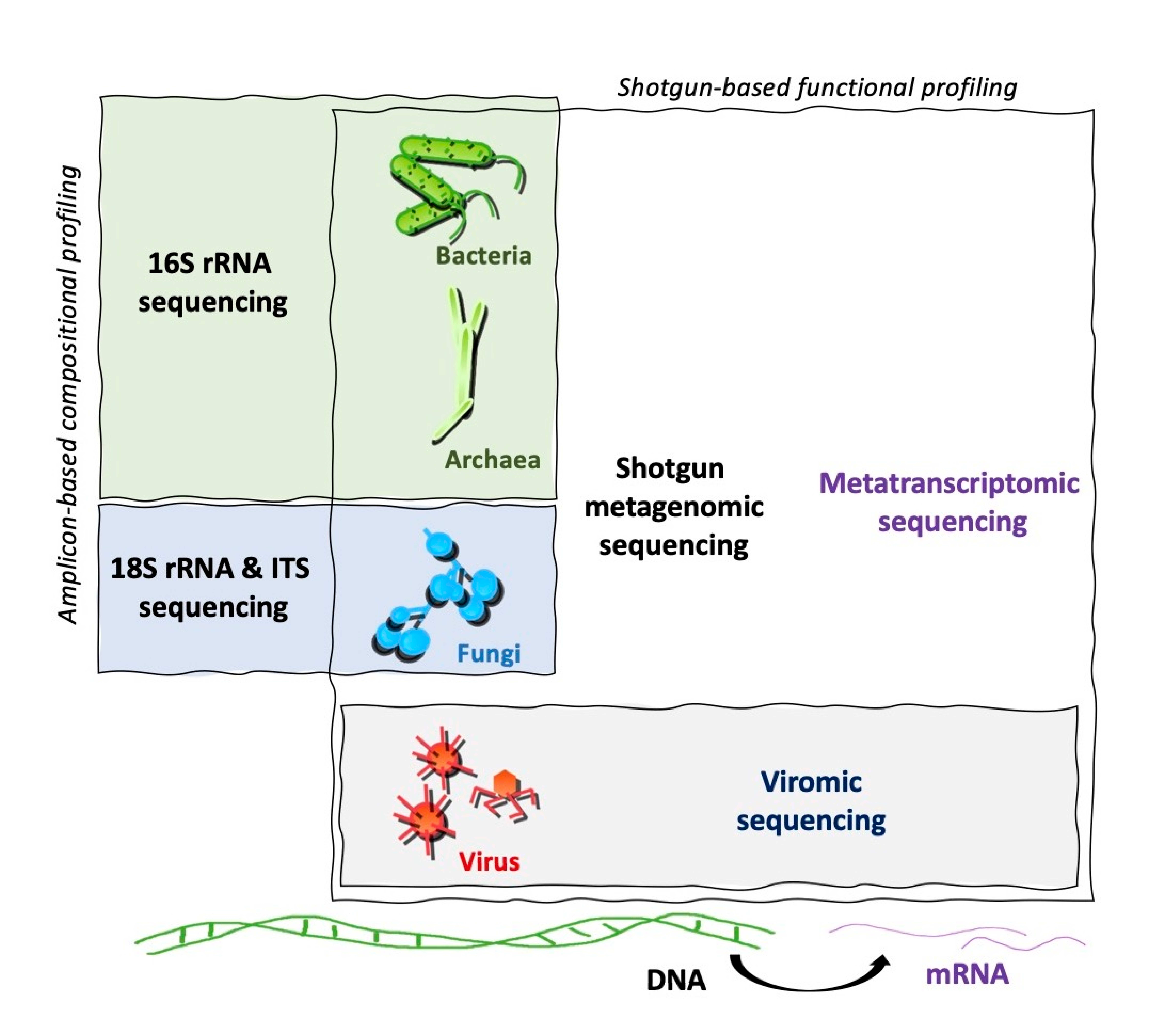
Biomolecules Free Full Text An Introduction To Next Generation Sequencing Bioinformatic Analysis In Gut Microbiome Studies Html
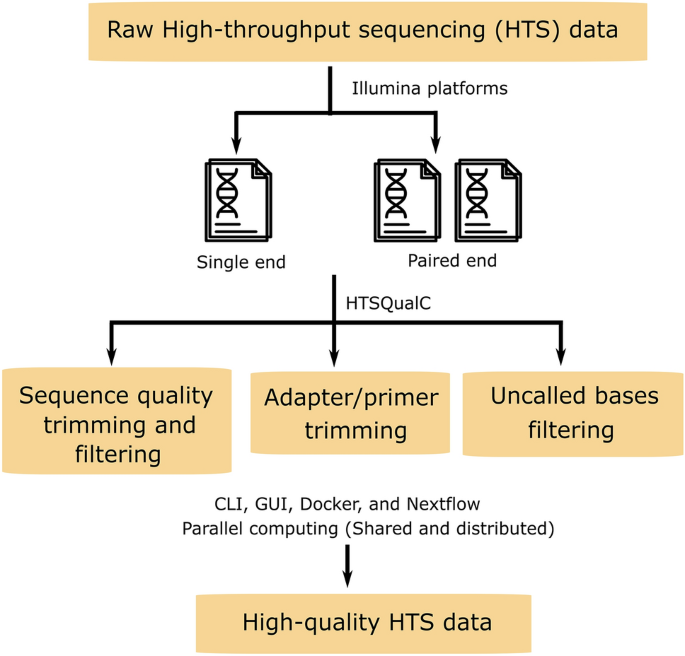
Htsqualc Is A Flexible And One Step Quality Control Software For High Throughput Sequencing Data Analysis Scientific Reports
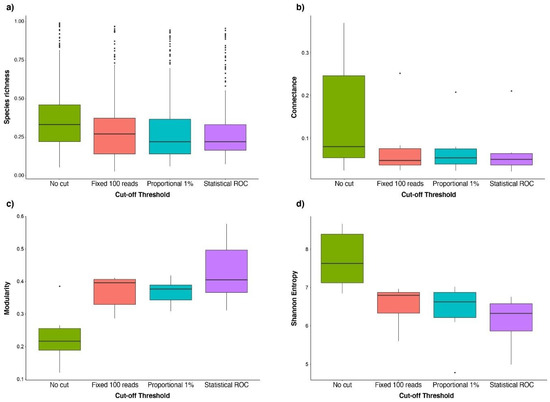
Diversity Free Full Text Harnessing The Power Of Metabarcoding In The Ecological Interpretation Of Plant Pollinator Dna Data Strategies And Consequences Of Filtering Approaches Html
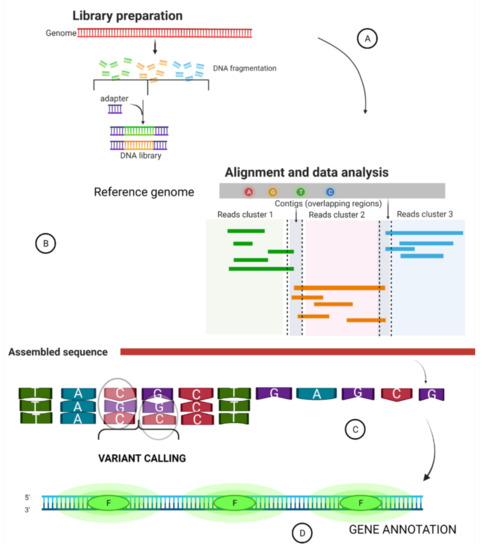
Biology Free Full Text Whole Genome Sequencing Contributions And Challenges In Disease Reduction Focused On Malaria Html
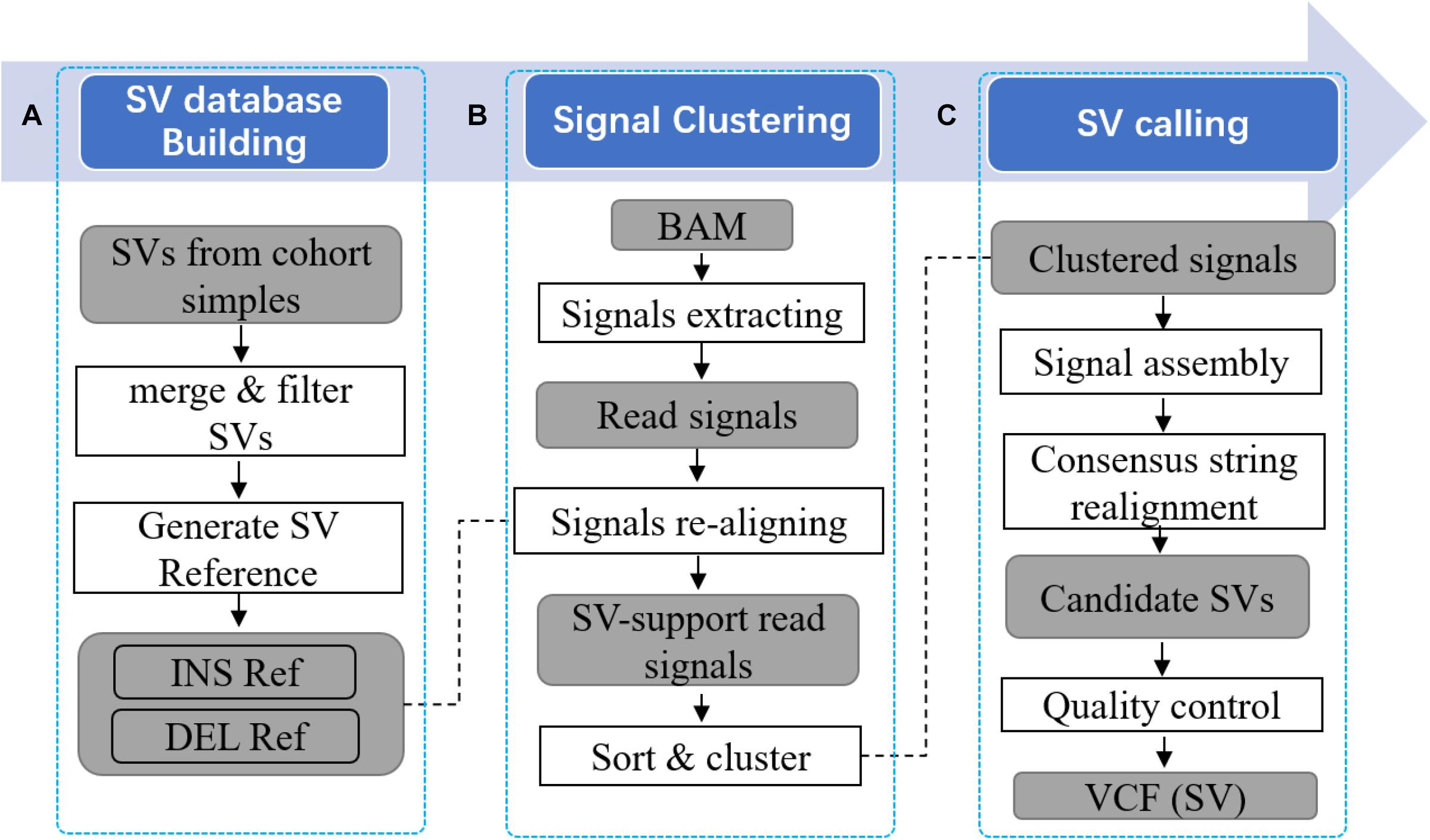
Frontiers Pansvr Pan Genome Augmented Short Read Realignment For Sensitive Detection Of Structural Variations Genetics

What Is Mate Pair Sequencing For

A Systematic Benchmark Of Nanopore Long Read Rna Sequencing For Transcript Level Analysis In Human Cell Lines Biorxiv
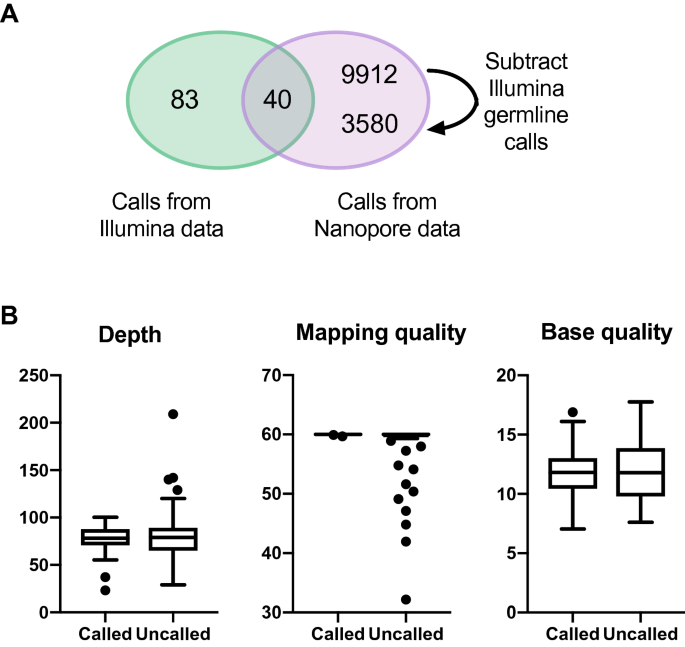
Short And Long Read Genome Sequencing Methodologies For Somatic Variant Detection Genomic Analysis Of A Patient With Diffuse Large B Cell Lymphoma Scientific Reports